
Note thatif an incorrectly incorporated nucleotide were removed by a proofreadingexonuclease (a 5’ to 3’ exonuclease in this hypothetical example), then theactivated end of the chain would be removed, and synthesis would stop.Īnswer 5.5. Chain synthesis would occur in a 3’ to 5’ direction. All these steps are similar to those inthe tail-growth mechanism at the 3’ end, except that the nonactivated end ofthe incoming nucleotide initiates the reaction with the activated end of thegrowing chain. The b- and g-phosphateswould be liberated as pyrophosphate. The 3’ hydroxylon an incoming nucleotide could react with the a-phosphate of the 5’ nucleotide by a nucleophilic attack. A hypothetical head-growth mechanismfor DNA synthesis would have the 5’ end of the primer at the active site this5’ end would have a triphosphate on the last nucletide added. 5.7 you would not expect to see a slow-sedimenting peakof nascent DNA.Īnswer5.4.
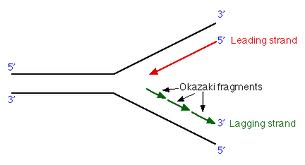

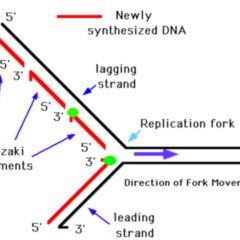
Therefore, by the model of semidiscontinuoussynthesis shown in Fig. Because the short Okazaki fragments shouldstill be in duplex with the large parental DNA strands, the duplex would notseparate from the bulk of the DNA. In an neutral sucrose gradient, the two strands ofthe DNA duplex should stay together. In contrast to the replication eyes, the two new strands are notsynthesized simultaneously at the replication fork in D loop replication.Īnswer 5.3. Theproduction of LL shows that replication is not random.Īnswer5.2. Thus, Rad59 promotes fork progression when Okazaki fragment processing is compromised and counteracts PCNA-K107 mediated cell cycle arrest.Answer 5.1. cdc9 rad59 double mutants did not alter PCNA ubiquitination but enhanced phosphorylation of the mediator of the replication checkpoint, Mrc1, indicative of increased replication fork stalling. To further understand how cells cope with nicks during replication, we utilized cdc9-1 in a genome-wide synthetic lethality screen and identified RAD59 as a strong negative interactor. Both enzymes reversed PCNA ubiquitination, arguing that the modification is likely triggered directly by nicks. To determine whether PCNA ubiquitination occurred in response to nicks or the lack of PCNA-DNA ligase interaction, we complemented cdc9 cells either with wild-type DNA ligase I or Chlorella virus ligase, the latter of which fails to interact with PCNA. In support of this notion, a pol30K107 mutation alleviated cell cycle arrest in cdc9 mutants. Most importantly, this signal is crucial to activate the S phase checkpoint, which promotes cell cycle arrest. The modification at K107 is catalyzed by the E2 variant Mms2 together with Ubc4 and the E3 ubiquitin ligase Rad5. cerevisiae as a model system, we uncovered a novel and conserved ubiquitination pathway that targets proliferating cell nuclear antigen (PCNA) at lysine 107 when DNA ligase I activity is inhibited. How cells monitor and suppress such accumulation of DNA damage that arises due to defective Okazaki fragment processing is unclear. An individual harboring DNA ligase I mutations exhibited growth retardation, sunlight sensitivity, severe immunosuppression and developed lymphoma, indicating a link between defects in Okazaki fragment maturation and cancer predisposition. In humans, approximately 30 million Okazaki fragments are synthesized during every S phase and require further processing prior to DNA ligation. DNA ligase I, encoded by the CDC9 gene in Saccharomyces cerevisiae, is an essential enzyme that catalyzes the ligation of newly synthesized DNA on the lagging strand called Okazaki fragments.


 0 kommentar(er)
0 kommentar(er)
